 (From left) Nam Hojeong, Professor of Electrical, Electronics and Computer Engineering; Seo Jiwon, Professor of Chemistry; Bae Daehun, Master’s student of Electrical, Electronics and Computer Engineering; Kim Minsang, PhD candidate of Chemistry. Provided by GIST
(From left) Nam Hojeong, Professor of Electrical, Electronics and Computer Engineering; Seo Jiwon, Professor of Chemistry; Bae Daehun, Master’s student of Electrical, Electronics and Computer Engineering; Kim Minsang, PhD candidate of Chemistry. Provided by GIST
Since the discovery of penicillin, antibiotics have revolutionized the treatment of infectious diseases. However, due to their misuse and overuse, multidrug-resistant bacteria (MDR), which possess resistance to multiple drugs through genetic mutations, have emerged and now pose a new threat to human health. In this context, a Korean research team has attracted attention by introducing a next-generation antibiotic development technology based on artificial intelligence (AI) that can rapidly identify new drug candidates specifically targeting resistant bacteria.
On July 23, the Gwangju Institute of Science and Technology (GIST) announced that a joint research team led by Nam Hojeong, Professor of Electrical, Electronics and Computer Engineering, and Seo Jiwon, Professor of Chemistry, has developed an AI model that analyzes the relationship between the genetic information of various bacteria and the activity of antimicrobial peptides, and proposes peptide-based antibiotic candidates specific to bacterial species.
This model learns the correlation data between the unique genetic information of each bacterial species and various antimicrobial peptides, enabling the selection of antimicrobial peptides optimized for the bacteria causing an infection. As a result, it is now possible to identify candidate treatments tailored to specific bacterial species for precision medicine, and to precisely target pathogens that have acquired resistance to existing antibiotics through genetic mutations.
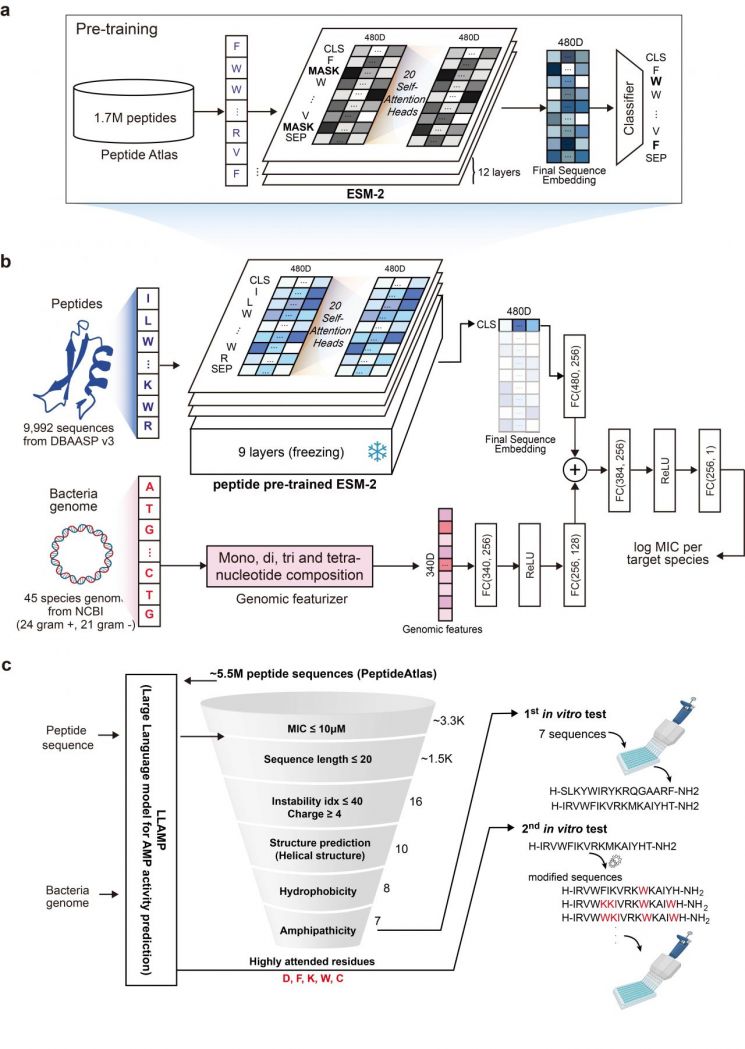
Until now, AI-based antimicrobial peptide research has been limited to simply predicting whether a peptide has antimicrobial activity, without considering the target bacterial species, which restricted its practical application. To overcome this, the research team developed 'LLAMP (Large Language model for AMP activity prediction),' the world's first AI model that learns from large-scale peptide data and utilizes the genome information of target bacterial species.
LLAMP predicts the minimum inhibitory concentration (MIC) of a peptide?an indicator of its activity against a specific bacterial species?by inputting the genome information of the bacteria and the peptide sequence. The model was developed by first training a language model pre-trained on protein data with large-scale peptide data to help it understand the unique 'language' of peptides, and then fine-tuning it based on combinations of bacterial genomes and peptides.
Compared to existing models, LLAMP improved the accuracy of antimicrobial activity prediction by at least 4% and up to 9%, and enhanced the prediction of activity values by at least 3% and up to 40%, demonstrating superior performance across all metrics.
The research team used LLAMP to screen approximately 5 million peptide sequences found in nature and, through attention analysis, identified specific amino acids that contribute to antimicrobial activity. Based on this analysis, they redesigned the peptides (sequence engineering) to further enhance their antimicrobial properties and confirmed their antimicrobial activity by directly applying them to pathogenic bacteria.
As a result, candidate substances screened using the genetic information of pathogens exhibited strong antimicrobial effects against highly toxic and resistant bacteria (ESKAPE), with minimum inhibitory concentration (MIC) values as low as 3.1 micromolar (μM). This experimentally demonstrated their high potential as actual therapeutic candidates.
Additionally, hemolytic toxicity tests were conducted on these peptide compounds using red blood cells, and they showed safety (hemolytic toxicity) and selectivity levels comparable to those of 'pexiganan,' a peptide antibiotic that has advanced to phase 3 clinical trials. Further research also revealed that highly active and selective antimicrobial peptides act by directly destroying bacterial cell membranes through the arrangement and amphipathic properties of specific amino acids derived from a helical structure.
The research team emphasized that the significance of this achievement lies in demonstrating that AI can go beyond simply mimicking existing drugs to analyze the genetic characteristics of pathogens and design new therapeutics optimized for them. In particular, this represents the introduction of a new drug development platform capable of tracking and responding to the evolution of antibiotic resistance in real time, which is expected to have a significant academic and industrial impact.
Nam Hojeong, Professor, stated, "The core of this research is the establishment of an AI-based drug development system that can rapidly propose antibiotic candidates based on the genetic information of newly emerging resistant bacteria. This differentiates our model from previous ones by enabling the identification of species-specific peptides and the development of antibiotics specialized for resistant bacteria."
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.
![Clutching a Stolen Dior Bag, Saying "I Hate Being Poor but Real"... The Grotesque Con of a "Human Knockoff" [Slate]](https://cwcontent.asiae.co.kr/asiaresize/183/2026021902243444107_1771435474.jpg)














