Attracting Attention as a Foundational Technology
for Enhancing Precision in New Drug Development
Ajou University announced on July 22 that a joint research team led by Professor Park Daechan from the Department of Molecular Science and Technology at the College and Graduate School of Advanced Bio-Convergence, Ajou University, and Professor Nam Jaehwan from the Department of Biomedical Science at the Catholic University of Korea, has developed a new technology capable of precisely quantifying the length of the poly(A) tail (polyadenylation tail), a core structure of mRNA therapeutics.
The results of this study were published online on June 28 in the renowned journal "Molecular Therapy," which specializes in cell and gene therapy as well as RNA-based therapeutics, under the title "3AIM-seq: Quality assessment of mRNA therapeutics using sequencing for 3′ poly(A) tails of in vitro-transcribed mRNA."
This research was co-supervised by Professor Park Daechan and Professor Nam Jaehwan as corresponding authors. Seo Jina, a master's graduate of Ajou University (currently at Nexai), Park Hyojeong, Ph.D. from the Catholic University of Korea (currently at the Korea Health Industry Development Institute, Disease Control Division), and Oh Ahyoung, a master's graduate of the Catholic University of Korea (currently at the Catholic University of Korea Convergence Science and Technology Research Institute), participated as co-first authors. The research teams of Professor Jang Hyesik from Seoul National University and Dr. Kim Cheona from the Korea Research Institute of Bioscience and Biotechnology also contributed as co-authors.
mRNA (messenger ribonucleic acid) functions in cells to produce proteins. mRNA therapeutics work by instructing cells to produce proteins that either compensate for deficiencies causing diseases or induce immune responses against specific diseases. mRNA technology is being widely researched for applications such as infectious disease vaccine development, cancer, genetic disorders, and autoimmune disease treatment. In fact, in response to the global spread of COVID-19, mRNA technology was used in the development of COVID-19 vaccines, leading to unprecedentedly rapid deployment and administration.
The poly(A) tail (polyadenylation tail), which the joint research team focused on, is a core structure of mRNA therapeutics. The poly(A) tail refers to a repeated adenine nucleotide sequence located at the 3' end of mRNA, playing a crucial role in maintaining mRNA stability and regulating protein expression efficiency. Therefore, it directly affects the efficacy and stability of mRNA drugs.
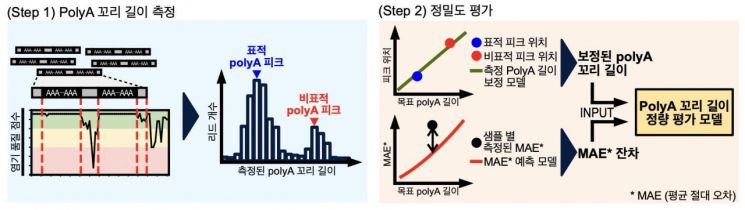
For the development and manufacturing process of mRNA therapeutics, it is essential to precisely measure and control the quality of the poly(A) tail's length and structure. However, because the poly(A) tail is a homopolymer structure composed of tens to hundreds of repeated identical adenine bases, conventional sequencing technologies frequently encounter insertion and deletion (indel) errors. This has limited the ability to quantitatively evaluate the poly(A) tail. In particular, to improve the uniformity of the poly(A) region in mRNA therapeutics, methods involving the insertion of short non-homopolymer linker sequences are increasingly being used, making structural analysis even more complex.
To address this, the joint research team designed and produced four types of in vitro-transcribed mRNA (IVT mRNA) with structures suitable for therapeutic use, as well as synthetic single-stranded DNA (ssDNA) oligonucleotides to serve as standards. Based on these, they developed a new sequencing analysis method called 3AIM-seq.
Focusing on the fact that accurate evaluation of poly(A) tail length and structure is directly linked to the stability and expression efficiency of mRNA therapeutics, the research team produced four types of in vitro-transcribed mRNA (IVT mRNA) with structural diversity, including the presence or absence of linker structures. These were then used to validate the performance of the 3AIM-seq analysis method.
Additionally, using the Illumina platform, a leading technology in next-generation sequencing, the team developed a bioinformatics algorithm capable of precisely calculating poly(A) tail length. This approach minimized errors that can occur during PCR amplification, enabled highly precise length prediction (with a precision of ±5 nt), and allowed for reliable identification of complex structures depending on the presence of linker insertions. By introducing a calibration model using the standard material, the research team improved analytical accuracy and succeeded in quantitatively assessing the heterogeneity of poly(A) tail length in actual in vitro-transcribed mRNA (IVT mRNA).
This study presents a new analysis method that offers both precision and reproducibility in structural quality assessment of mRNA therapeutics, demonstrating its potential as a practical tool for simultaneously evaluating the length and structure of the poly(A) tail during therapeutic development and manufacturing. As a result, it is expected to contribute to the establishment of quality control standards and regulatory compliance for a wide range of mRNA-based therapeutics, including vaccines and anticancer agents.
Professor Park Daechan of Ajou University stated, "This research outcome is a new analytical technology that enables quantitative evaluation of the 'poly(A)' structure, which has been considered the most challenging aspect of mRNA therapeutics to analyze." He added, "The new sequencing analysis method (3AIM-seq) developed by our team can serve as a key tool for quality control of mRNA vaccines and therapeutics in the future."
In this joint study, Professor Park Daechan's team at Ajou University was responsible for developing next-generation sequencing methods and experimental designs optimized for poly(A) tail analysis, as well as the 3AIM-seq algorithm. Next-generation sequencing (NGS) is an analytical technique that enables high-throughput, rapid, and accurate reading of DNA or RNA sequences compared to conventional methods, and is widely used in various fields such as biomarker research and gene expression studies. Professor Nam Jaehwan's team at the Catholic University of Korea was responsible for designing and producing in vitro-transcribed mRNA (IVT mRNA) with structures suitable for therapeutic use.
This research was supported by the Ministry of Education's G-LAMP project, the Ministry of Science and ICT's Mid-Career Researcher Program, and the Ministry of Food and Drug Safety.
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.
![Clutching a Stolen Dior Bag, Saying "I Hate Being Poor but Real"... The Grotesque Con of a "Human Knockoff" [Slate]](https://cwcontent.asiae.co.kr/asiaresize/183/2026021902243444107_1771435474.jpg)














