Professor Kim Bitnaeri and Research Team at Seoul National University Uncover Principles of RNA-Protein Interactions
 From the left, Bitnari Kim, Head of RNA Research Group (co-corresponding author), Jongwoo Bae, Researcher (first author), Jongseo Kim, Research Fellow (co-corresponding author). In the back is the mass spectrometer used in this study (Orbitrap Fusion Lumos, Thermo Fisher Scientific). Photo by Institute for Basic Science (IBS)
From the left, Bitnari Kim, Head of RNA Research Group (co-corresponding author), Jongwoo Bae, Researcher (first author), Jongseo Kim, Research Fellow (co-corresponding author). In the back is the mass spectrometer used in this study (Orbitrap Fusion Lumos, Thermo Fisher Scientific). Photo by Institute for Basic Science (IBS)
[Asia Economy Reporter Kim Bong-su] Last year, Korean researchers including Professor Kim Bit-naeri of Seoul National University, who gained attention for creating the world's first high-resolution genetic map of the COVID-19 virus, have now found the key to unlocking the secrets of RNA-protein interactions essential for regulating life phenomena.
The Institute for Basic Science (IBS) announced on the 25th that the research team led by Research Fellow Kim Jong-seo (Assistant Professor at Seoul National University) and Director Kim Bit-naeri (Distinguished Professor at Seoul National University) of the RNA Research Group developed a technique to precisely identify ‘RNA binding sites’ on intracellular proteins according to the type of nucleotide. This is expected to greatly contribute to elucidating the principles of RNA-protein interactions essential for regulating life phenomena.
Identifying RNA binding sites is a crucial step in understanding cellular function regulation. Proteins bind to RNA to regulate protein production processes such as translation efficiency, stability, and intracellular localization, thereby expressing normal genes. To confirm RNA binding sites, a ‘mass spectrometry’ method is required, which measures the mass of small protein fragments and infers the amino acid sequence and position within the protein.
Last year, the research team developed a technique to find RNA binding sites by applying short-wavelength ultraviolet light (UVC) and hydrofluoric acid to form cross-links between uracil in RNA and proteins. However, this technique made it difficult to comprehensively understand RNA-protein interactions because the cross-linking efficiency was very low and only one type of RNA base, uracil, could be used.
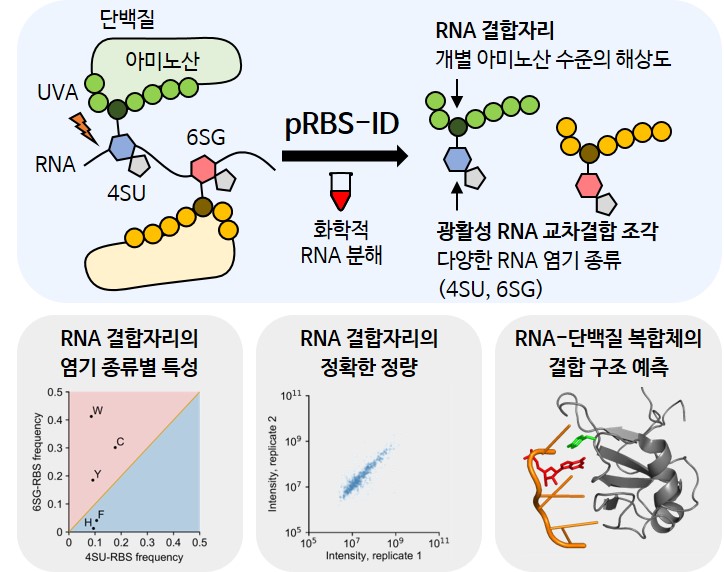 Schematic diagram of pRBS-ID research using photoactivatable RNA.
Schematic diagram of pRBS-ID research using photoactivatable RNA.For pRBS-ID, photoactivatable RNA-protein covalent bonds are first formed using UVA. Then, proteins are converted into peptides, and RNA is chemically degraded using hydrofluoric acid. Subsequent mass spectrometry allows high-resolution identification of RNA binding sites at the individual amino acid level based on the mass of photoactivatable RNA fragments crosslinked to peptides. In this study utilizing pRBS-ID, the researchers characterized the properties of RNA binding sites according to nucleotide types, accurately quantified RNA binding sites, and established a foundation for analyzing RNA-protein interactions based on structural data of RNA-protein complexes.
The research team overcame the limitations of last year’s study by using long-wavelength ultraviolet light (UVA) and photoactivatable RNA labeling. First, intracellular RNA was labeled with two types of photoactivatable nucleotides: 4-thiouridine and 6-thioguanosine. Then, UVA was used to induce specific cross-linking with bound proteins, and the long RNA bound to proteins was chemically hydrolyzed by hydrofluoric acid treatment. The molecular weight of the remaining RNA fragments was then confirmed by mass spectrometry. Based on this information, they efficiently identified specific RNA binding sites corresponding to each nucleotide type of 4-thiouridine and 6-thioguanosine. Additionally, they improved data analysis software to accurately quantify peptides containing RNA binding sites.
Thus, the research team closely elucidated differences in RNA-protein interaction patterns according to nucleotide types and cross-linking methods. This achievement encompasses not only newly discovered photoactivatable RNA binding sites but also previously identified RNA binding sites using UVC. Furthermore, by integrating RNA-protein complex structural data, they confirmed that the RNA binding sites identified by mass spectrometry correspond to nucleotide types and interactions consistent with known protein-RNA structures. They also revealed RNA-protein interactions that could not be detected with existing data due to protein structural instability through RNA binding sites.
The research team stated, "The mass spectrometry technique developed this time and the information on more than 3,000 newly identified RNA binding sites can be utilized to understand the regulation of life phenomena by RNA-protein interactions." They added, "In particular, combining metabolic labeling of photoactivatable RNA with recently dramatically improved protein structure prediction programs will enable detailed understanding of the dynamic changes and molecular mechanisms of RNA-protein interactions."
The research results were published online on the 15th in the international journal Nature Communications (IF 14.92).
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.
![Clutching a Stolen Dior Bag, Saying "I Hate Being Poor but Real"... The Grotesque Con of a "Human Knockoff" [Slate]](https://cwcontent.asiae.co.kr/asiaresize/183/2026021902243444107_1771435474.jpg)















