Professors Park Man-sung and Kim Yoon-ki of Korea University, and Professor Baek Dae-hyun of Seoul National University Joint Research Team
Also Mapping Patterns Within the Human Body... Expected to Aid Vaccine and Therapeutic Development
[Asia Economy Reporter Kim Bong-su] Domestic researchers have created a high-resolution map measuring the pattern of COVID-19 virus infection and spread in the human body, and based on this, identified key factors controlling the virus's spread. This is expected to aid vaccine and treatment development.
The National Research Foundation of Korea announced on the 25th that a joint research team including Professors Park Man-sung and Kim Yoon-ki from Korea University and Professor Baek Dae-hyun from Seoul National University produced a high-resolution map measuring the patterns of the translatome and transcriptome over time after infection by SARS-CoV-2, the virus causing COVID-19. The transcriptome refers to the total RNA transcribed from the genome containing genetic information, and the translatome refers to the comprehensive pattern of the translation process that decodes the transcriptome to produce proteins.
To develop vaccines and treatments for COVID-19, it is essential to understand how the virus infects and spreads within the human body through self-replication. In other words, understanding the pathophysiology and elucidating the principles of viral gene expression are necessary, and observing changes in gene expression patterns of both human and virus after infection is essential.
SARS-CoV-2 has 12 genes containing information on characteristic structural proteins such as the spike protein and replication proteins used to spread the genome within the host. The transcription process that produces messenger RNA (mRNA), an intermediate step in protein production from these genes, is relatively well known. However, the translation process, where proteins are generated from mRNA, is not well understood. There has also been a lack of data measuring changes in host and viral genome expression over time after infection, making it difficult to understand the pathological mechanisms.
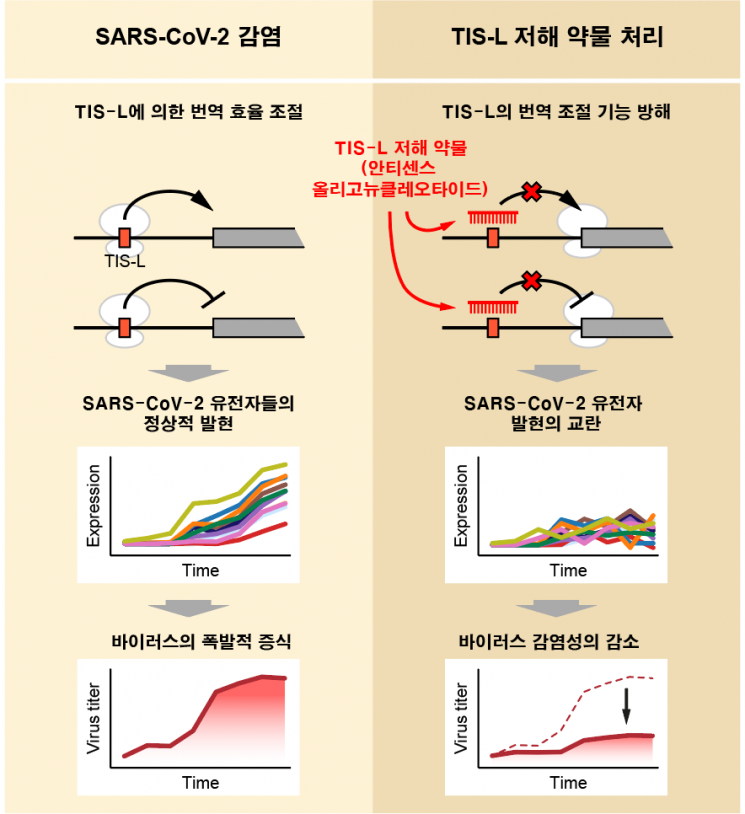
The research team succeeded in measuring the translation and transcription patterns of human cells and viral genomes at various time points after SARS-CoV-2 infection and obtaining large-scale next-generation sequencing (NGS) data, a technique for analyzing DNA or RNA sequences on a large scale and at high speed. Based on the SARS-CoV-2 translatome map obtained, they identified a factor regulating the efficiency of viral protein production, which they named TIS-L (translation initiation site located in the leader). The team experimentally verified that TIS-L significantly affects the translation efficiency of viral proteins, including the spike protein, a major target of COVID-19 vaccines.
By analyzing changes in human gene expression patterns, they also detected groups of genes showing similar expression patterns over time after viral infection. They confirmed that genes related to cellular stress respond strongly in the early stages of infection, while genes related to immune response react significantly in the later stages.
The research team stated, "This study is expected to help understand the infection mechanism and pathophysiology of SARS-CoV-2 and provide clues for therapeutic research targeting TIS-L." The study results were published on the 25th in the international journal Nature Communications.
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.
![Clutching a Stolen Dior Bag, Saying "I Hate Being Poor but Real"... The Grotesque Con of a "Human Knockoff" [Slate]](https://cwcontent.asiae.co.kr/asiaresize/183/2026021902243444107_1771435474.jpg)















