KAIST Chemistry Department Research Team
[Asia Economy Reporter Kim Bong-su] A domestic research team has developed a model that can identify new drug candidate substances more than twice as fast as existing methods using artificial intelligence.
KAIST announced on the 17th that Professor Kim Woo-yeon’s research team in the Department of Chemistry developed a protein-ligand interaction prediction model with higher generalization performance than existing methods by integrating physicochemical ideas into AI deep learning.
A ligand refers to a substance that specifically binds to large biomolecules such as receptors and plays a crucial role in the body as well as in drug development. To discover drug candidate molecules, it is important to find ligands that strongly bind to target proteins. However, conducting exhaustive experiments on millions to tens of millions of random ligand libraries to find effective substances requires astronomical time and cost. To reduce this time and cost, virtual screening technology based on protein-ligand interaction prediction has recently attracted attention.
Existing AI models for interaction prediction show high predictive performance for structures used in training but suffer from over-fitting, resulting in low performance for new protein structures. Over-fitting typically occurs when the amount of data is insufficient relative to the model’s complexity. This study focused on solving the over-fitting problem to develop a prediction model that shows consistent performance across various proteins.
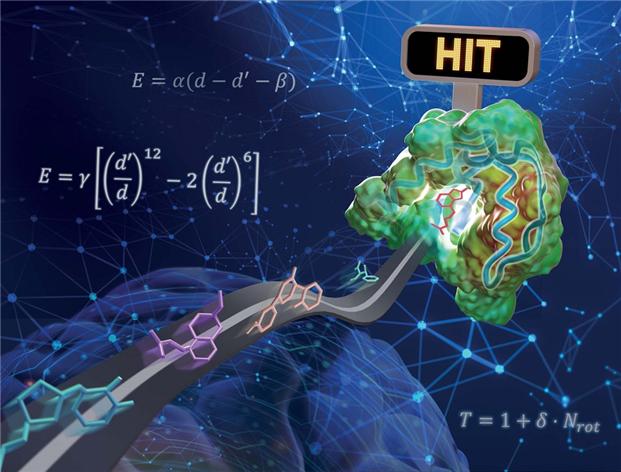
The research team applied physicochemical ideas to the deep learning model to reduce model complexity while addressing the over-fitting problem by augmenting insufficient data through physical simulations. They modeled van der Waals forces and hydrogen bonding between protein atoms and ligand atoms using physicochemical equations and predicted parameters through deep learning, enabling predictions that satisfy physical laws.
They also noted that the protein-ligand crystal structures used for training are the most stable structures experimentally determined. To augment the insufficient experimental data, they generated hundreds of thousands of artificial data consisting of unstable protein-ligand structures and used them for training. As a result, the model was trained to predict actual structures more stably compared to the generated structures.
To verify the performance of the developed model, the research team used the `CASF-2016 benchmark' as a control group. This benchmark includes essential tasks in the drug development process, such as docking to find structures close to experimentally determined crystal structures among various protein-ligand structures and screening to find protein-ligand pairs with relatively strong binding affinity. The verification test results showed higher docking and screening success rates compared to previously reported technologies, with screening performance approximately twice as high as the best previously reported performance.
Another advantage of the physics-based deep learning methodology developed by the research team is that the prediction results are physically interpretable. This is because the final interaction values are predicted through physicochemical equations optimized by deep learning. By analyzing the contribution of interaction energy per atom within the ligand molecule, it is possible to identify which functional groups played an important role in protein-ligand binding. Such information can be directly utilized to enhance performance through future drug design.
A research team official said, "Generalization issues have always been emphasized as important problems in chemistry and bio fields where data is scarce," adding, "The physics-based deep learning methodology used in this study can be applied not only to protein-ligand interaction prediction but also to various physical problems."
The results of this study were published as a cover paper and selected as the 'Pick of the Week' in the April 13, 2022 issue of the international journal 'Chemical Science (IF=9.825)'.
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.