Institute for Basic Science Kim Bitnaeri RNA Research Team Leader: "Likely to Contribute to Therapeutic Development"
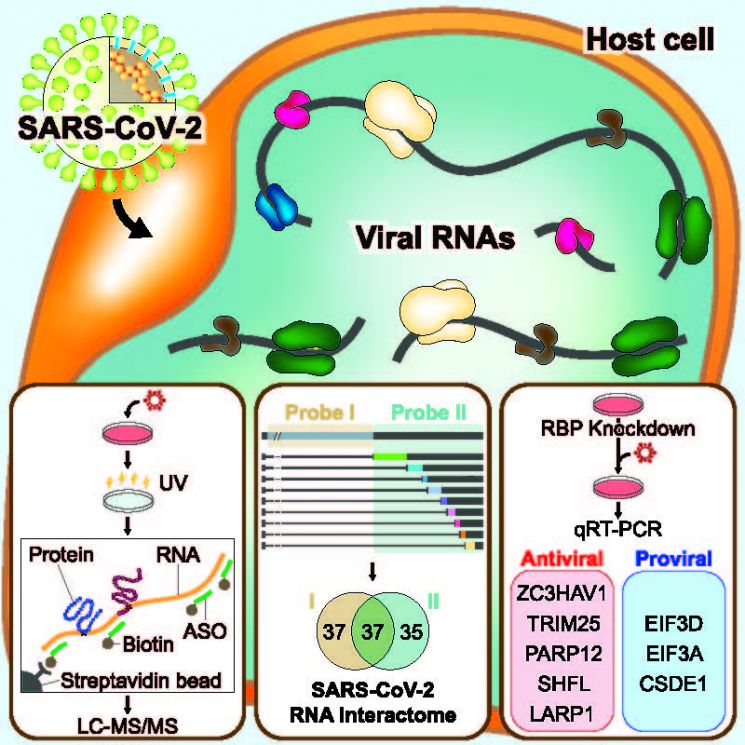 Schematic diagram of coronavirus replication inhibition mechanism. Image courtesy of the Institute for Basic Science.
Schematic diagram of coronavirus replication inhibition mechanism. Image courtesy of the Institute for Basic Science.
[Asia Economy Reporter Kim Bong-su] Domestic researchers have discovered proteins that control the replication of the COVID-19 virus, which is expected to contribute to the development of treatments.
The Institute for Basic Science (IBS) announced on the 28th that the research team led by Kim Bit-naeri, head of the RNA Research Group and professor at the Department of Biological Sciences at Seoul National University, discovered proteins that directly bind to the coronavirus RNA and regulate its replication. With this research, the team completed a high-resolution proteome map following last year's high-resolution genetic map of SARS-CoV-2, the cause of COVID-19. IBS stated that this is expected to contribute to the development of COVID-19 therapeutics.
Coronaviruses are a type of RNA virus. They invade host cells and produce and translate their genomic RNA, which contains their genetic information, to create various non-structural proteins. These non-structural proteins block the host cell's primary immune response (innate immunity) and replicate the viral genome. Subsequently, subgenomic RNA is produced from the genomic RNA. This subgenomic RNA serves as a blueprint for various structural proteins (such as spike and envelope proteins) that make up the virus. Structural proteins and genomic RNA assemble virus particles, which exit the cell to infect new cells.
Thus, proteins in the host cell that bind to genomic RNA and subgenomic RNA play a crucial role in coronavirus replication. However, little has been known about these proteins until now.
The research team developed a technology to isolate and identify only proteins that bind to specific RNA to find proteins that specifically bind to SARS-CoV-2. Using this, they identified all 109 proteins that bind to SARS-CoV-2 RNA. Among these, 37 were confirmed to bind commonly to both genomic RNA and subgenomic RNA.
They also conducted comparative analysis with HCoV-OC43, another type of coronavirus. They classified proteins that act commonly across the coronavirus family and those that bind exclusively to SARS-CoV-2, analyzing their respective functions. As a result, they discovered eight proteins that aid viral replication and 17 antiviral proteins. They elucidated not only all proteins that directly bind to SARS-CoV-2 but also their effects on viral replication.
Furthermore, through cross-analysis based on RNA big data, they completed a network map between host cells and SARS-CoV-2. Based on understanding protein molecular interactions centered on viral RNA, they revealed part of the complex relationship between host cells and the virus. For example, host cell proteins LARP1 and SHIFTLESS inhibit viral protein production, thereby blocking viral replication.
The research team stated, "This study has deepened our understanding of coronavirus replication," and added, "It is also expected to open possibilities for developing antiviral drugs targeting proteins that directly bind to SARS-CoV-2."
The research results were published online on the 27th (Korean time) in the international journal Molecular Cell (IF 15.584).
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.