IBS Professor Kim Bitnaeri's Research Team
Discovers Method to Stabilize RNA and Increase Protein Production
"Can Be Used to Enhance mRNA Therapeutic Performance"
Domestic researchers have developed a technology that can increase RNA stability and protein production in viruses. It is explained that this can be used to enhance the performance of gene therapies such as messenger ribonucleic acid (RNA) vaccines, which have been widely used in the fight against the COVID-19 virus.
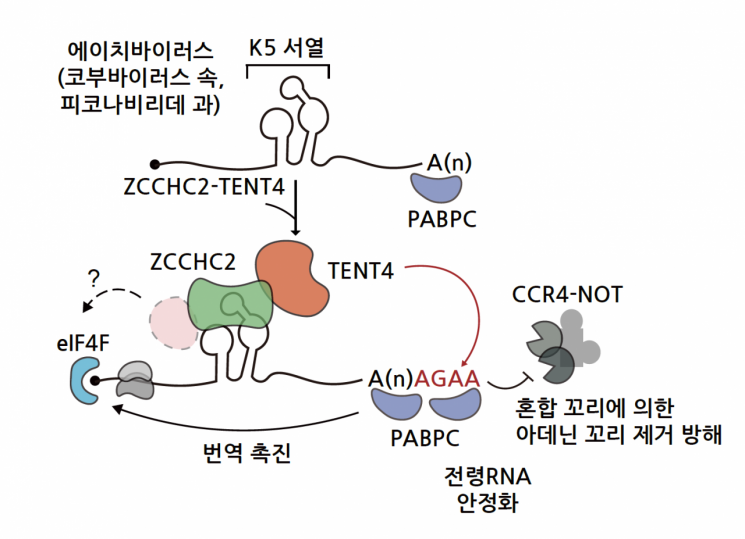 Mechanism of action of the K5 sequence identified through RNA screening. Image source: Provided by IBS
Mechanism of action of the K5 sequence identified through RNA screening. Image source: Provided by IBS
The Institute for Basic Science (IBS) announced on the 6th that the research team led by Kim Bitnaeri, head of the RNA Research Center (Distinguished Professor at the Department of Biological Sciences, Seoul National University), discovered RNA sequences that increase RNA stability and protein production in viruses by using a high-throughput sequencing technology capable of simultaneously analyzing hundreds of viral RNAs. This sequence, named K5’, is expected to significantly enhance the performance of RNA therapeutics.
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease, the demand for virus research has exploded to overcome infectious diseases. Viruses have evolved various biological mechanisms to rapidly adapt to their environment. Although research on viruses has greatly contributed to the advancement of life sciences, it has so far been limited mainly to a very small number of viruses known to be medically or industrially important. However, because viruses have very diverse types and life cycles, they may represent an unexplored “treasure trove of knowledge” from which new insights about genes and RNA can be obtained.
To find regulatory sequences that contribute to RNA stabilization and protein production in viruses, the research team collected RNA sequence information from all viruses known to infect humans. They then applied unbiased selection criteria to represent all viruses and selected 143 representative viral sequences. These sequences were cut into fragments of the same length (130 nucleotides), creating about 30,000 fragments, which were introduced into cells to analyze whether each viral sequence could increase RNA stability and protein production.
Through this screening process, the research team was able to identify multiple regulatory sequences that increase RNA stabilization and protein production, and identified 16 sequences that contribute to both RNA stabilization and protein production. Among them, the most effective sequence was named ‘K5’ and was analyzed in detail. K5 is a sequence located near the 3’ end of Aichivirus. Aichivirus belongs to the genus Kobuvirus, family Piconaviridae, and is a virus with a single-stranded RNA genome. Although it is widespread worldwide, it causes only mild gastroenteritis and has been scarcely studied due to its low pathogenicity.
The research team revealed that the K5 regulatory sequence helps RNA avoid degradation and produce more protein by generating a mixed tail on the RNA using the TENT4 protein, which is part of the ZCCHC2-TENT4 protein complex. K5 forms two hairpin structures, and when the ZCCHC2-TENT4 protein complex binds to the K5 sequence, the TENT4 protein forms a mixed tail on the adenine tail of the virus. This mixed tail slows down RNA degradation, thereby increasing RNA stability.
Furthermore, the clinical applicability of the K5 sequence was confirmed through experiments inserting the K5 sequence into viral vectors used in gene therapy, resulting in a significant increase in gene delivery efficiency of the viral vectors. Additionally, when inserted into mRNA vaccines, it was confirmed that the mRNA was stabilized and produced large amounts of protein for an extended period. This demonstrates that the K5 sequence can be utilized to enhance the performance of RNA therapeutics.
Director Kim said, “The K5 sequence of viral RNA increases RNA stability and protein production. Using the K5 sequence, the stability and performance of mRNA vaccines and gene therapies can be improved.” He added, “This achievement is difficult to accomplish with the conventional approach that focuses only on highly pathogenic viruses.” He also emphasized the significance of the research by stating, “Even mild viruses can evolve into serious viruses in the future, so it is important to study a diverse range of viruses without bias.”
This research achievement was published online on the 6th in Cell (IF 66.850), a prestigious journal in the field of biology.
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.