KAIST Research Team Develops New Matrix Decomposition Technique
Achieves First Identification of Principles Behind Inter-Chromosomal Interactions
Domestic researchers have unveiled the three-dimensional genome structure within the cell nucleus related to cancer and aging.
The Korea Advanced Institute of Science and Technology (KAIST) announced on the 10th that Professor In-Kyung Jung's research team from the Department of Biotechnology, in collaboration with Professor Yong-Dae Shin's research team from the Department of Mechanical Engineering at Seoul National University and Professor Jung-Mo Choi's research team from Pusan National University, discovered new principles for the formation of the 3D genome structure within the cell nucleus and identified mediators that regulate it.
Research on the 3D genome structure has revealed that the genome inside the cell nucleus is organized hierarchically, with each structure involved in regulating various gene expressions. It has recently been shown that the 3D genome structure is closely related to disease-specific gene expression in complex diseases such as cancer and aging. However, most studies have been limited to interactions within chromosomes, which are relatively easier to observe. Due to limitations in experimental observation techniques, research on inter-chromosomal interactions on a larger scale has been scarcely conducted.
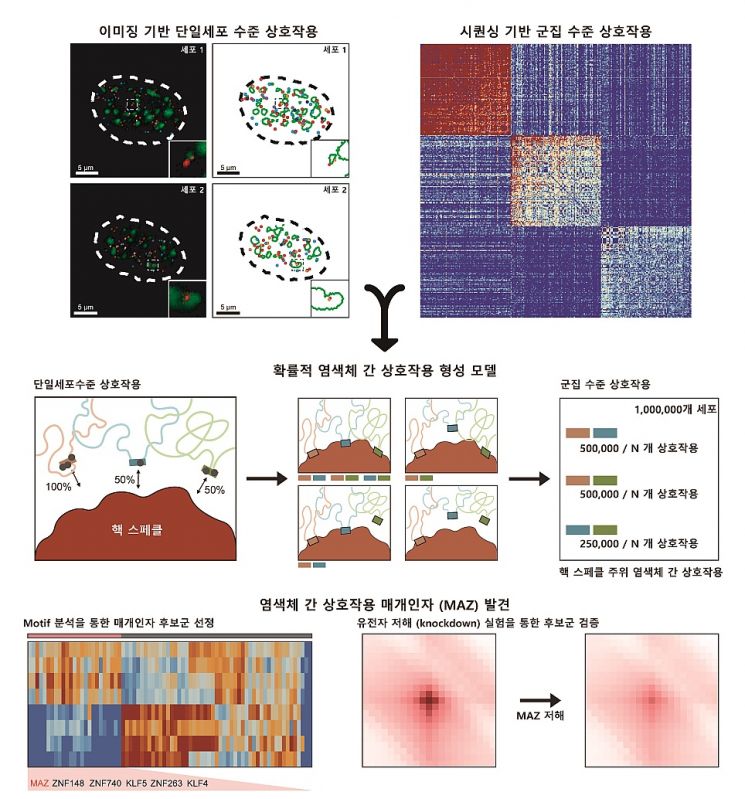
The research team developed a novel machine learning algorithm that effectively extracts inter-chromosomal interaction information from genome 3D structure data by utilizing a matrix decomposition analysis technique, and validated it through DNA imaging methods.
Using this analytical algorithm, the team extracted and analyzed inter-chromosomal interaction information from various cell lines and observed that inter-chromosomal interactions located around nuclear speckles (membrane-less structures within the nucleus) are commonly conserved across multiple cells. Furthermore, through DNA motif analysis, they discovered for the first time that the inter-chromosomal interactions around speckles are mediated by the MAZ protein. They also found that inter-chromosomal interactions occur differently in each cell at the single-cell level. The team proposed that, unlike previously known fixed interactions, inter-chromosomal interactions are probabilistically determined through individual interactions between nuclear bodies and genome regions, thereby elucidating the principles of inter-chromosomal interactions for the first time.
The results of this study were published on the 5th in the international journal Nucleic Acids Research (IF=19.16) under the title: Probabilistic establishment of speckle-associated inter-chromosomal interactions.
This study is significant as it revealed the previously unknown formation principles of inter-chromosomal interactions and the role of the MAZ protein as a mediator, providing fundamental insights into the larger-scale 3D genome structure.
Jae-Geon Joo, an integrated MS-PhD student at KAIST who led the research, explained, "This study uncovered the formation principles of inter-chromosomal interactions that had been obscured due to limitations in experimental techniques." Professor Jung also stated, "This achievement is expected to enhance understanding of the importance of interactions between nuclear bodies and the genome in gene expression regulation according to 3D genome structure and in identifying causes of chromosomal abnormalities frequently reported in cancer and other diseases."
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.