[Asia Economy Yeongnam Reporting Headquarters Reporter Dongyeol Hwang] A research team at Ulsan National Institute of Science and Technology (UNIST) has developed software to analyze chromatin immunoprecipitation (ChIP), which is used to investigate the binding sites of specific proteins.
Chromatin immunoprecipitation (ChIP) is widely used to investigate the binding sites of specific proteins, and the latest experimental technique using exonuclease, called ChIP-exo, can identify binding sites with high resolution.
However, the task of distinguishing peaks, which are the actual binding sites of the target protein on DNA, requires a labor-intensive additional verification step by researchers, limiting the ability to process large volumes of data quickly and accurately.
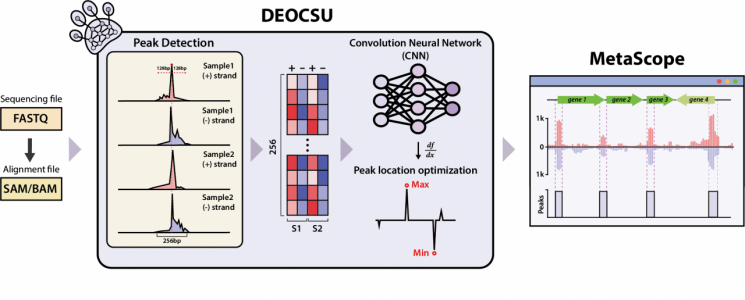
To address this issue, Professor Donghyuk Kim’s team from the Department of Energy and Chemical Engineering at UNIST developed DEOCSU (DEep-learning Optimized ChIP-exo peak calling SUite), a deep learning-based ChIP-exo peak selection software.
DEOCSU first detects peak candidates from ChIP-exo data aligned to a reference sequence. Each detected signal is converted into image data, and then a convolutional neural network, a technique that divides the image into small units and analyzes each part using trained data, is used to select the actual peaks.
Each selected peak can be used to estimate position optimization and binding size. The resulting data can be checked using MetaScope, a visualization software developed in-house.
The DEOCSU model trained on ChIP-exo data from the Escherichia coli K-12 MG1655 strain accurately selected peaks not only for the data used in training but also for unknown ChIP-exo data.
Professor Donghyuk Kim of the Department of Energy and Chemical Engineering said, “Despite the useful advantage of identifying protein-DNA interactions at high resolution, the use of ChIP-exo experimental technology has been limited due to analysis difficulties. With the development of DEOCSU, the burden on researchers for analysis can be overcome, accelerating the progress of related research.”
This research was conducted with support from the Ministry of Science and ICT’s Bio·Medical Technology Development Project and the Donggurami Foundation’s Innovative Science and Technology Center·Program Competition Project.
The research results were published on January 25 in Briefings in Bioinformatics, a leading journal in bioinformatics research.
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.
![Clutching a Stolen Dior Bag, Saying "I Hate Being Poor but Real"... The Grotesque Con of a "Human Knockoff" [Slate]](https://cwcontent.asiae.co.kr/asiaresize/183/2026021902243444107_1771435474.jpg)















