 ▲Superbacteria
▲Superbacteria
[Asia Economy Reporter Junho Hwang] Domestic researchers have discovered genes resistant to antibiotics in the water of the Han River. These genes are among those that make up bacteriophages, viruses that parasitize bacteria. They function similarly to antibiotic resistance genes found in super bacteria but are new genes. The research team expects this to broaden the horizon of antibiotic resistance gene research.
The National Research Foundation of Korea announced on the 8th that Professor Jangcheon Cho and Dr. Gira Moon from Inha University, Professor Sanghee Lee from Myongji University, and Professor Changjun Cha from Chung-Ang University secured genes causing antibiotic resistance from bacteriophages present in the Han River and named them "Han River Virome Beta-lactamase (HRV)" and others.
Antibiotic Resistance Genes Float in Han River Water
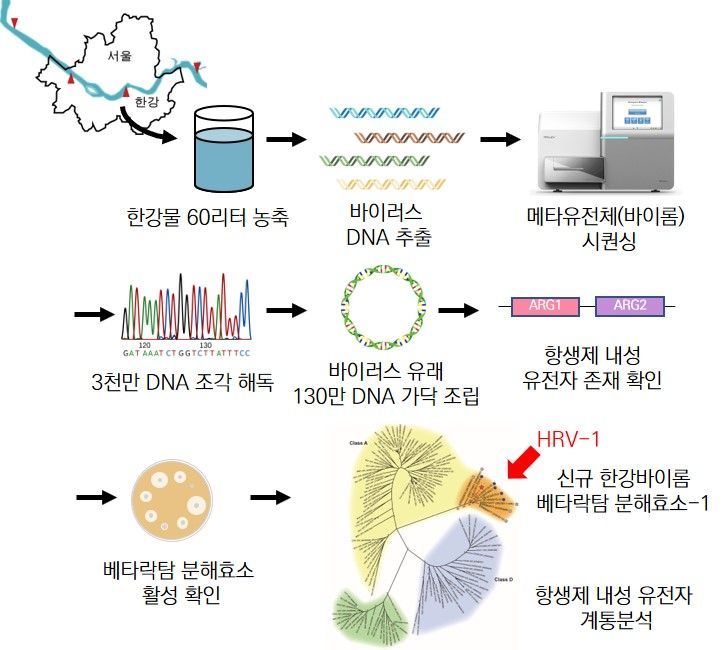 Schematic Diagram of Han River Virus Genome Exploration Containing Multiple Antibiotic Resistance Genes
Schematic Diagram of Han River Virus Genome Exploration Containing Multiple Antibiotic Resistance Genes
The research team found antibiotic resistance genes in the Han River that are resistant to beta-lactam antibiotics such as penicillin, cephalosporin, and carbapenem, which are found in pathogenic bacteria. Beta-lactam antibiotics are the most widely used antibiotics, and among them, carbapenem is known as a strong antibiotic and the last line of defense in treating pathogens.
The team collected 10 liters of surface water from six points along the Han River, removed bacteria, and then concentrated only viruses. They extracted nucleic acids and obtained 1.3 million sequence fragments, identifying 25 antibiotic resistance genes among them. Four of these phage-derived genes were inserted into Escherichia coli to verify whether they produce functional enzymes. As a result, E. coli carrying these genes showed up to 16 times higher antibiotic resistance compared to the control group.
The research team stated that this is the first antibiotic resistance gene with proven function found in viruses existing in the natural environment. Although the core nucleotide sequences are identical to beta-lactamase genes (antibiotic resistance genes) found in existing super bacteria, the rest differ, leading the team to conclude that these are new genes and to assign them names.
Whether It Spreads to Bacteria Is a Follow-up Study
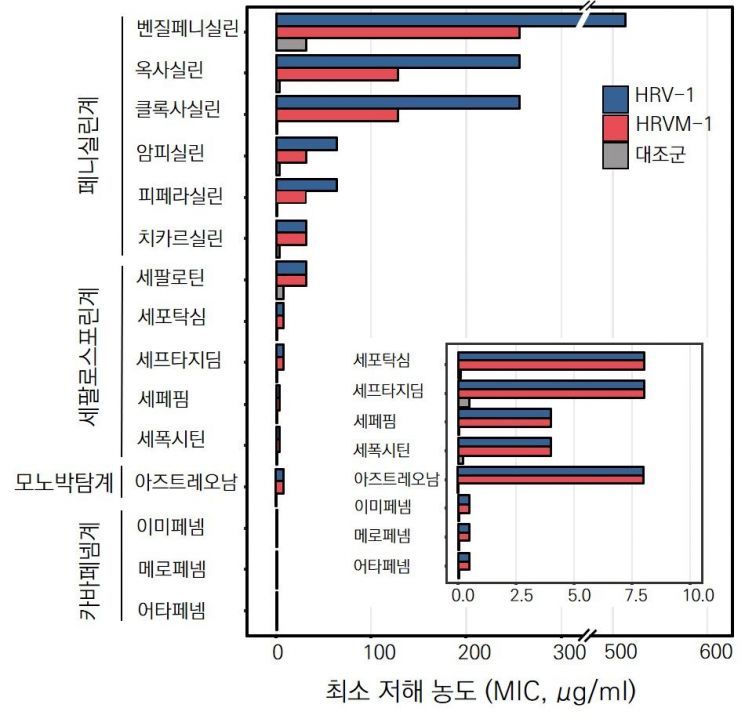 This is the result of determining the minimum inhibitory concentration through Escherichia coli expression of a novel beta-lactamase.
This is the result of determining the minimum inhibitory concentration through Escherichia coli expression of a novel beta-lactamase.
This study overturns previous findings that antibiotic resistance genes found in the natural environment are rare and mostly non-functional. However, the actual impact of these genes on humans remains a future task.
The research team stated, "These genes do not infect human cells through Han River water," and added, "It is a future research task to determine why these genes show antibiotic resistance, how they are transmitted to which bacteria via bacteriophages, what actions they perform inside bacteria, and whether they spread between bacteria."
They further emphasized, "Since environments with relatively high antibiotic exposure such as lungs, wastewater treatment plants, and fish farms are expected to harbor more diverse virus-derived antibiotic resistance genes, we plan to expand the 'large-scale viral metagenomic research' used in this study to various environments."
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.