Development of 'ChIP-mini' technology to analyze DNA-protein binding sites using 5,000 times fewer cells
Enables precise identification of pathogenic gene regulation mechanisms in Salmonella
Lays the foundation for large-scale data production in biofoundry research
In elucidating the pathogenic expression mechanisms of bacteria or in biofoundry technology, techniques that identify and analyze the binding sites of proteins attached to DNA are crucial. A new technology has been developed that can accurately separate binding sites using significantly smaller sample amounts than before.
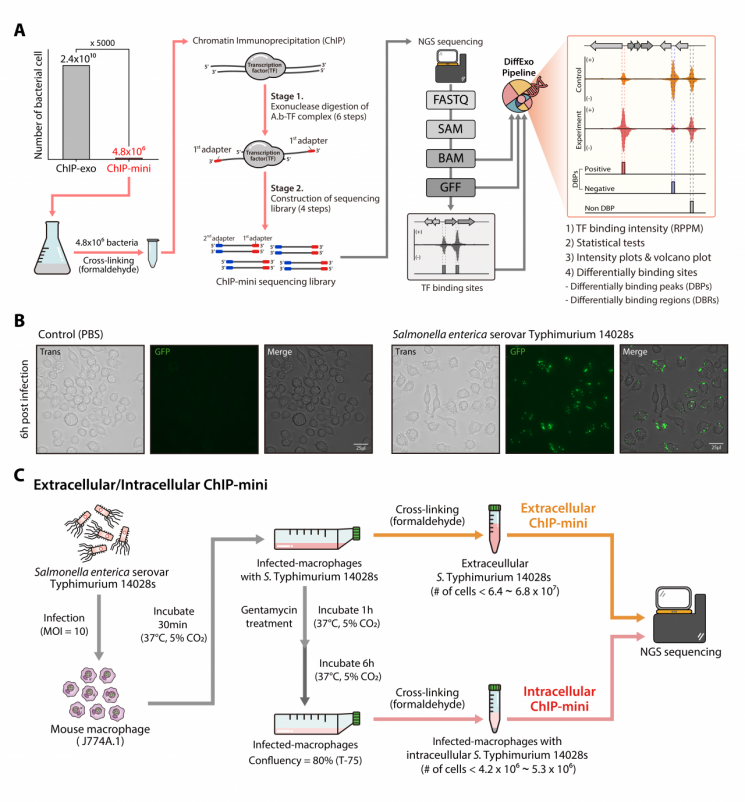
Professor Donghyuk Kim’s team from the Department of Energy and Chemical Engineering at UNIST and Professor Eunjin Lee’s team from the Department of Life Sciences at Korea University have developed a ‘mini’ chromatin immunoprecipitation method (ChIP-mini) that can analyze the binding sites of special proteins on DNA at high resolution using 5,000 times fewer cells than existing methods.
DNA, which stores genetic information, is composed of long chains of nucleotide molecules. Cells regulate gene expression by binding special proteins called transcription factors to specific sites on the DNA.
Chromatin immunoprecipitation (ChIP) is a technique that isolates only these special proteins bound to DNA and identifies the DNA fragments to which they are attached. The newly developed ‘ChIP-mini’ technology can separate binding sites with a precision of one base pair (approximately 0.34 nm) using only 4.8 million cells, about 5,000 times fewer than conventional methods.
Thanks to this, even when multiple proteins bind in close proximity, each binding site can be accurately distinguished and analyzed. To achieve a similar level of precision with the latest ChIP-exo experimental method, approximately 10 billion to 100 billion cells were required.
The collaborative research team demonstrated the performance of ChIP-mini by isolating an extremely small amount of Salmonella bacteria that caused infection in the host and quantitatively analyzing changes in the DNA binding locations and intensities of two special proteins (H-NS and RpoD) inside the bacteria.
Salmonella strongly binds the H-NS protein to DNA outside the host macrophages to suppress the expression of pathogenic genes. However, once it successfully enters the host cells, the binding strength of H-NS protein decreases, and the RpoD protein binds to selectively activate pathogenic genes. This
is a mechanism to regulate pathogenicity outside the host cells to avoid immune attacks from the host.
Since Salmonella infects host cells in extremely small amounts, it was difficult to perform such analysis with existing ChIP methods due to insufficient numbers of Salmonella cells inside the host cells. For quantitative analysis, the research team used their self-developed statistical program, DiffExo.
Additionally, the unit cost per analysis using the ChIP-mini technology is about 20,000 KRW, which is more than 12 times lower than before.
 Identification of Salmonella DNA-binding protein binding sites inside and outside macrophages using an optimized ChIP-mini experimental method.
Identification of Salmonella DNA-binding protein binding sites inside and outside macrophages using an optimized ChIP-mini experimental method.
Junyoung Park, a co-first author and a PhD candidate at UNIST, explained, “By combining the developed technology with an NGS automation platform, we can gain an advantage in producing large-scale binding data required for biofoundry development quickly and cheaply.” He added, “We are currently conducting additional research to ensure compatibility with next-generation sequencing (NGS) automated instruments.”
Biofoundry technology involves operating microorganisms like bacteria as if they were semiconductor foundries to produce high value-added proteins and other products. For microbial gene editing and optimal circuit design, large amounts of binding data are necessary.
Professor Donghyuk Kim stated, “This research is valuable as a foundational technology in the biofoundry field, including elucidating gene expression networks of infectious microorganisms and discovering bio-parts.”
This study was published on February 10 in the international journal Nucleic Acids Research. The research was supported by the Ministry of Science and ICT’s Bio and Medical Technology Development program and the National Research Foundation’s Basic Research Laboratory support project.
© The Asia Business Daily(www.asiae.co.kr). All rights reserved.













